Features
Features
Genetic copy number variation (CNV) analysis of schizophrenia
- ツイート
- 2018/05/11
Graduate School of Medicine / Institute of Advanced Research
Designated Assistant Professor Itaru Kushima
Source: IAR Letter, vol.16, March 2018
*about IAR: Nagoya University's Institute for Advanced Research was established to produce internationally recognized academic research of the highest caliber, and to contribute to society through the research achievements at the Institute.
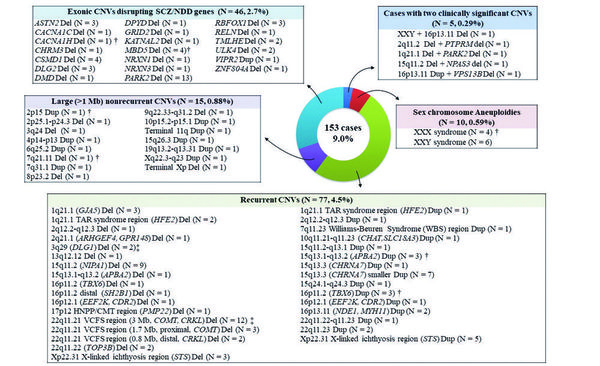 Figure shows clinically significant (or pathogenic) copy number variations (CNVs) identified in patients with schizophrenia. 158 CNVs were detected in 153 of 1699 patients (9.0%). Five patients had two clinically significant CNVs. The CNVs were classified into four categories: 1) sex chromosome aneuploidies, 2) recurrent CNVs, 3) large nonrecurrent CNVs, and 4) exonic CNV disrupting SCZ/NDD genes. Number and % frequency of each category are presented in parentheses. † indicates that de novo CNV was detected in one patient, and ‡ indicates that de novo CNV was detected in two patients. Abbreviations: Del: deletion; Dup: duplication; NDD: neurodevelopmental disorder; SCZ: schizophrenia
Figure shows clinically significant (or pathogenic) copy number variations (CNVs) identified in patients with schizophrenia. 158 CNVs were detected in 153 of 1699 patients (9.0%). Five patients had two clinically significant CNVs. The CNVs were classified into four categories: 1) sex chromosome aneuploidies, 2) recurrent CNVs, 3) large nonrecurrent CNVs, and 4) exonic CNV disrupting SCZ/NDD genes. Number and % frequency of each category are presented in parentheses. † indicates that de novo CNV was detected in one patient, and ‡ indicates that de novo CNV was detected in two patients. Abbreviations: Del: deletion; Dup: duplication; NDD: neurodevelopmental disorder; SCZ: schizophrenia
Genetic copy number variation (CNV) contributes substantially to human evolution, normal phenotypic variation, and human disease. To date, thousands of different genomic duplications and deletions, each spanning hundreds to millions of base pairs, have been mapped genome-wide, and collectively account for a significant fraction of human genetic variation. The importance of several rare CNVs (<1% in population) in the risk of schizophrenia has been suggested in previous studies. We performed a high-resolution genome-wide CNV analysis of 1699 schizophrenia patients and 824 healthy controls. Our study revealed high genetic heterogeneity of schizophrenia and its clinical features and raises the possibility that genomic instability is involved in its pathogenesis, which may be related to the increased burden of de novo CNVs and the variable expressivity of CNVs.
INTRODUCTION
Human genomes vary from one to another in many ways, and the totality of this genetic variation underlies the heritability of human traits and genetic diseases. Recent genome studies have revealed that various types of genetic variations are present in the human genome. Among them, copy number variations (CNVs) are a particularly important component of genetic variations, affecting a greater fraction of the genome. CNVs are defined as copy number changes including deletions and duplications of genomic regions that range from one kilobase (kb) to megabases (Mbs) in size. Recently, several rare CNVs (<1% in population) have been found to be associated with a risk of schizophrenia. Schizophrenia is a major psychiatric disorder that affects about 1% of the population. This disorder is characterized by psychotic symptoms (delusions and hallucinations), negative symptoms (social withdrawal and blunted affect), and cognitive impairments. Although the precise pathogenesis of schizophrenia remains largely unclear, epidemiological studies have shown that genetic factors have an important role in the risk of this disorder, with heritability estimated at 70–90%. Eleven large rare CNVs have been reported to be associated with the risk of schizophrenia with odds ratios (ORs) of 2 to over 50. Such CNVs included deletions at 1q21.1, NRXN1, 3q29, 15q11.2, 15q13.3, and 22q11.2 and duplications at 1q21.1, 7q11.23, 15q11.2-q13.1, 16p13.1 and 16p11.2. These CNVs were also implicated in the risk for other neurodevelopmental and psychiatric disorders (variable expressivity of CNVs). Despite the progress, small CNVs (<100 kb) have not been fully studied in schizophrenia. Furthermore, the pathogenesis of this disorder has not been elucidated. Thus, we performed a high-resolution genome-wide CNV analysis to address these issues.
CNV analysis
Using a microarray technology called array comparative genomic hybridization, we performed a high-resolution genome-wide CNV analysis of 1699 schizophrenia patients and 824 healthy controls (1). This was the largest CNV study for a psychiatric disorder in Japan. Overall, we identified 7066 rare (<1%) CNVs and 70.0% of them were small (<100 kb). These small CNVs were difficult to detect in previous studies. We examined the genome-wide CNV burden in schizophrenia and confirmed the previous findings that patients with schizophrenia had a greater genome-wide burden of rare exonic CNV than healthy controls.
We then looked into clinically significant (or pathogenic) CNVs, which were previously associated with the risk of neurodevelopmental or psychiatric disorders (e.g., intellectual disability, autism spectrum disorder, attention deficit hyperactivity disorder, bipolar disorder). Such CNVs were identified in 9.0% (153/1699) of patients (Figure) and 3.2% (26/824) of healthy controls. The excess of these CNVs in patients was statistically significant (odds ratio = 3.04, P = 9.3 × 10-9), supporting the pathogenicity of such CNVs in schizophrenia. This result also indicated that schizophrenia is a genetically heterogeneous disorder and that genetic risk factors are shared among different neurodevelopmental and psychiatric disorders. For individual CNVs, we obtained evidence for a significant association of X-chromosome aneuploidies (XXX/XXY) and 22q11.2 deletions with schizophrenia. The 22q11.2 deletions have been reported as the most important genetic risk factor for schizophrenia in previous studies.
Phenotypic analysis
To characterize clinical features of patients with clinically significant CNVs, we examined their clinical information in detail. As a result, 41.7% of patients with such CNVs had a history of congenital phenotypes (e.g., congenital heart defects) or premorbid developmental problems (e.g., intellectual disability). In addition, the rate of treatment resistance to antipsychotics (primary medication for schizophrenia) was significantly higher in patients with these CNVs than in those without them (36.1% vs 16.9%, odds ratio = 2.79, P = 0.0036). This indicated that CNV findings may be useful in predicting the response to antipsychotics in patients with schizophrenia. We also found more severe clinical manifestations in patients with two clinically significant CNVs.
Biological pathways
Identification of biological pathways is critical for understanding its pathogenesis and development of novel treatment. For that purpose, we performed bioinformatic analysis (i.e., gene set analysis) using our CNV datasets. As a result, we identified multiple biological pathways implicated in the pathogenesis of this disorder: oxidative stress response, genomic integrity, gene expression regulation, cell adhesion, neurotrophin signaling, kinase, synapse, small GTPase signaling and endocytosis. The importance of synapse, cell adhesion, and small GTPase signaling in schizophrenia were strongly supported by previous genetic studies. Novel biological pathways included oxidative stress response and genomic integrity. Oxidative stress, which is an imbalance between reactive oxygen species and the antioxidant defense system, induces DNA damage such as double-strand breaks. Although increased oxidative stress has been reported in these patients, its role in the pathogenesis remained unclear until now. Genomic integrity is essential for neuronal survival and normal neuronal function. This pathway included 'DNA repair', 'DNA replication' and 'DNA recombination', errors which are involved in the major mechanisms of CNV formation. Therefore, CNVs in patients which affect the oxidative stress response or genomic integrity may promote genomic instability that underlies high de novo CNV rates and a greater burden of rare CNVs in schizophrenia. In addition, genomic instability may influence the genome of somatic cells (neurons) and increase somatic mutations. Consistent with this, an increased copy number of L1 retrotransposon in neurons has been recently reported in schizophrenia. Somatic mutations (mosaicism) are also implicated in variable expressivity of CNVs. Finally, a genetic model of schizophrenia is provided in Figure.
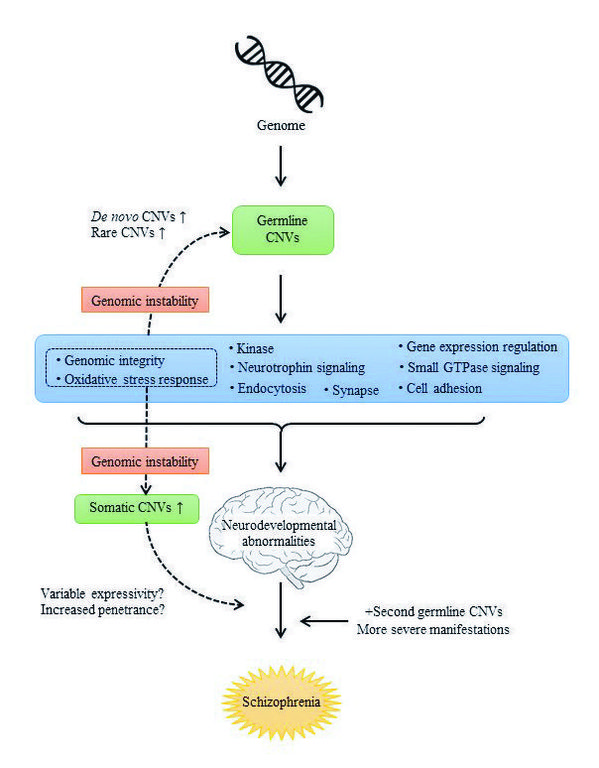 A genetic model of schizophrenia
A genetic model of schizophrenia
In pathogenesis of this disorder, nine pathways (oxidative stress response, genomic integrity, gene expression regulation, cell adhesion, neurotrophin signaling, kinase, synapse, small GTPase signaling, and endocytosis) are affected by CNVs. Disruption of genomic integrity and oxidative stress response induces genomic instability, which is involved in germline CNV formation and somatic CNV formation in neurons. The former account for an increased rate of de novo or rare CNVs and the latter for the variable expressivity of CNVs.
Research Summary
We revealed the high genetic heterogeneity of schizophrenia and clinical features of patients with pathogenic CNVs, and raised the possibility that genomic instability is involved in its pathogenesis, which may be related to the increased burden of de novo CNVs and the variable expressivity of CNVs.
References
(1) High-resolution copy number variation analysis of schizophrenia in Japan. Kushima I, Aleksic B, Nakatochi M, Shimamura T, Shiino T, Yoshimi A, Kimura H, Takasaki Y, Wang C, Xing J, Ishizuka K, Oya-Ito T, Nakamura Y, Arioka Y, Maeda T, Yamamoto M, Yoshida M, Noma H, Hamada S, Morikawa M, Uno Y, Okada T, Iidaka T, Iritani S, Yamamoto T, Miyashita M, Kobori A, Arai M, Itokawa M, Cheng MC, Chuang YA, Chen CH, Suzuki M, Takahashi T, Hashimoto R, Yamamori H, Yasuda Y, Watanabe Y, Nunokawa A, Someya T, Ikeda M, Toyota T, Yoshikawa T, Numata S, Ohmori T, Kunimoto S, Mori D, Iwata N, Ozaki N. Mol Psychiatry, 2017 Mar; 22(3):430-440.
 |
Itaru KUSHIMA : Designated Assistant Professor Graduate School of Medicine / Institute of Advanced Research, Nagoya University |
Related Links
NU Research
(English)

