Nano-sized springs that always twist in the same direction may help bridge the gap between nanoscale and macroscale movement
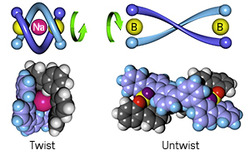
Fig. 1: Schematic illustration showing how the addition and removal of a sodium ion (Na+) causes the molecular spring, consisting of two oligomers (blue) bridged by boron (B) atoms, to twist and untwist repeatedly in only one direction.
Adapted from Ref. 1 c 2010 E. Yashima
Most biological functions are driven by 'molecular machines', and developing synthetic devices that mimic their behavior could pave the way for numerous technological applications. Scientists have already constructed a variety of nano-sized machines, including molecular muscles, motors and shuttles, but it remains difficult to exercise fine control over their movements using external stimuli such as chemicals or light.
Synthetic helical molecules called 'molecular springs' have been proposed as a method of translating motion from the nano- to the macroscale. The main challenge in using these components is how to control the contraction (twisting) and extension (untwisting) of the springs. Some helical molecules that can extend and contract have been reported recently, but it has so far proved impossible to ensure that twisting only occurs in one direction.
Now, Kazuhiro Miwa, Yoshio Furusho and Eiji Yashima from Nagoya University's GCOE for the Elucidation and Design of Materials and Molecular Functions1 have succeeded in constructing a double-stranded helix molecule that consistently twists in a clockwise direction when exposed to sodium ions and then untwists in the opposite direction when released. "Unidirectional twisting is one of the essential motions for transforming energy into mechanical work, as observed in a number of biological systems," says Yashima.
The molecule designed by the research team is made up of two parallel oligomer (short polymer) strands bridged by two boron atoms (Fig. 1). The molecule has a natural twist, or chirality, with the boron atoms and oligomers meeting at each end of the bridge to form 'spiroborate' functional groups. When sodium ions are added to the system, they lock onto the spiroborate groups, causing the oligomers to twist up around the ion to form a double helix-like structure. To untwist the helix, a special selective ligand is added to the system to snatch the sodium away from the oligomer. Once the sodium is removed, the helix unwinds and returns to its original form.
The researchers showed that the twisted spring is less than half the length of the untwisted form. What's more, they observed the same behavior over many cycles of adding and removing the sodium ion. "The unidirectional spring-like motion can be repeated many times with the helix retaining its inherent chirality," Yashima explains.
"To translate the spring-like unidirectional motion into macrocyclic motion, we now need to incorporate the molecule into bulk systems, such as liquid crystals and organogels," says Yashima. "One could also foresee associating the extension/contraction motion with a catalytic function in some mechanochemical reactions."
Affiliated Researchers
The Nagoya University affiliated researchers mentioned in this highlight are from the Elucidation and Design of Materials and Molecular Functions GCOE program of the Department of Chemistry.
Reference
- Miwa, K., Furusho, Y. & Yashima, E. Ion-triggered spring-like motion of a double helicate accompanied by anisotropic twisting. Nature Chemistry 2, 444-449 (2010). | article