A variety of protein structural interactions act to control the activity of actin filaments
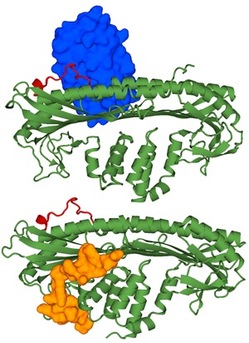
Fig. 1: Crystal structures of the CP/V-1 complex (top) and the CP/CARMIL complex (bottom). CP is shown in green, the CP α-tentacle in red, V-1 in blue, and CARMIL in orange.
Actin is a ubiquitous filament protein that plays an essential role in numerous biological processes, including muscle contraction, neural development and the division and movement of cells. It is regulated by actin capping protein (CP), which binds tightly the ends of actin filaments.
CP itself is inhibited by two proteins called V-1 and CARMIL, and until recently scientists were unsure of the exact molecular mechanisms responsible for this inhibition. Shuichi Takeda and colleagues at Nagoya University have now determined exactly how these three proteins interact, showing that V-1 and CARMIL inhibit CP by two distinct mechanisms1.
Takeda and his team used X-ray crystallography to examine molecules of CP bound to fragments of V-1 and CARMIL. In a previous study, they found that three particular amino acid residues, which constitute part of a structure called the 'α-tentacle', are essential for binding to the ends of actin molecules. In their latest analysis of the CP/V-1 complex (Fig. 1), they revealed that these α-tentacle residues also participate in V-1 binding.
V-1 inhibits CP function by a steric mechanism, meaning that it simply gets in the way of the binding sites. To illustrate this point, the researchers showed that when V-1 was overexpressed in cultured cells, it enhanced the elongation of actin filaments. This effect occurs because the V-1 prevents CP from binding to the filaments.
Unexpectedly, the analysis also revealed that the CP molecule actually consists of two domains, and that it undergoes conformational changes, with the two domains constantly twisting against each other.
On examining CARMIL, the researchers unveiled a very different inhibition mechanism to V-1. CARMIL binds to CP on the opposite end from the actin binding site, suggesting that it inhibits CP by an allosteric, or indirect, mechanism. CARMIL binds CP at a site that straddles the two domains, and inhibits CP by preventing the twisting movements that are essential for tight actin filament capping.
This is the first study to show that CP is not a rigid molecule but one that actually undergoes dynamic structural changes. CARMIL utilizes this intrinsic fluctuation of CP to suppress capping activity, and V-1 and CARMIL act together to regulate the assembly of actin filaments within the cell by fine-tuning CP activity.
"We are now interested in capturing the twisting motion of CP directly," says Takeda, "because we believe this intrinsic fluctuation is a key to understanding the actin filament capping process. We are therefore planning to investigate CP by a more dynamic method like nuclear magnetic resonance spectroscopy."
Affiliated Researchers
The Nagoya University affiliated researchers mentioned in this highlight are from the Advanced Systems Biology: Designing the Biological Function GCOE program
Reference
Takeda, S. et al. Two distinct mechanisms for actin capping protein regulation -- steric and allosteric inhibition. PLoS Biology 8: e1000416 (2010). | article