The protein fragments that cause allergic reactions could be identified using new bioinformatics software
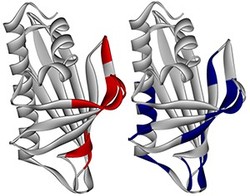
Fig. 1: The regions of birch pollen allergen that produce peaks on the AUF plot at low hydrophobicity (right, blue) correspond closely with the regions of the epitopes (red, left).
Mild allergies such as hay fever are very common in humans, and allergic reactions also have a role in conditions such as asthma. An allergic reaction is triggered when an environmental substance, or allergen, enters the body and a part of it called the epitope is recognized by antibodies produced by the immune system.
Now, a collaboration of researchers led by Shigeki Mitaku of the Nagoya University Center for Computational Science has developed a simple bioinformatics method for identifying protein fragments, such as epitopes, that undergo structural change upon protein-protein interaction1.
The researchers analyzed the amino acid sequences of 663 allergens and 539 non-allergens, and identified nearly 8,700 protein fragments that were unique to the allergens. The amino acids in these allergen unique fragments (AUFs) were found to have a characteristic distribution, with charged residues such as glutamic acid and lysine at the center flanked by a valley, whereas aromatic residues were found to have the inverse pattern.
Mitaku and his colleagues developed an 'AUF plot', a novel method for visualizing the distribution of amino acids in the protein fragments. An AUF plot is particularly useful for finding AUF-like fragments in an amino acid sequence that are located within loops in the moleccule, or at the ends of structural motifs called α-helices and β-pleated sheets.
To better understand how the AUF plot relates to epitope structure, the researchers examined birch pollen -- the only allergen with a well-characterized molecular structure -- both on its own and bound to the antibody that recognizes it. Analysis of the AUF plot revealed that the peaks correlated well to regions on the surface of the birch pollen allergen molecule that undergo structural changes when bound to an antibody (Fig. 1).
The researchers also noted that the size of the peak on the AUF plot corresponds to the extent to which the protein fragment has the characteristics of an AUF. This means that the method could be useful for predicting the protein fragments that undergo structural changes, such as the epitopes that are unique to allergens.
"Software that accurately predicts epitopes, either alone or in combination with experimental analyses, could be very useful," says Mitaku. "It could, for example, be used to assess the safety of genetically modified food products."
Affiliated Researchers
The Nagoya University affiliated researchers mentioned in this highlight are from the Center for Computational Science.
Reference
- Asakawa, N., Sakiyama, N., Teshima, R. & Mitaku, S. Characteristic amino acid distribution around segments unique to allergens. J. Biochem. 147, 127-133 (2010). | article