Solving the structure of the enzyme that allows some plants to make chlorophyll in the dark sheds light on photosynthesis
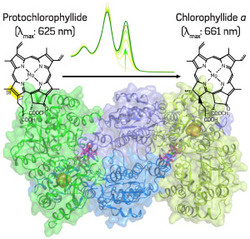
Fig. 1: Protochlorophyllide reduction and the X-ray crystal structure of DPOR NB protein. Pchlide (top left) is converted to chlorophyllide a (top right), the direct precursor for chlorophyll a, by DPOR (bottom center). The conversion can be observed by a spectral change (top center).
c 2010 N. Muraki, J. Nomata, G. Kurisu, Y. Fujita
A team led by Yuichi Fujita of the Nagoya University GCOE for Systems Biology has successfully solved the structure of a key enzyme involved in the biosynthesis of chlorophyll in the photosynthetic bacterium Rhodobacter capsulatus1.
Chlorophylls are pigments essential for photosynthesis, the process by which photosynthetic organisms use the energy of sunlight to synthesize carbon compounds beginning with carbon dioxide and water. "Elucidating the complex chemical pathways underlying the biosynthesis of chlorophyll is essential for understanding the evolution of photosynthesis and should help produce better crop plants," says Fujita.
The ability of chlorophyll pigments to trap light energy efficiently depends on the absorption properties of what is known as a 'tetrapyrrole ring system'. Maturation of this central ring system occurs at the penultimate stage of its synthesis when a chemical called protochlorophyllide (Pchlide) is converted into the direct precursor of chlorophyll a by the 'Pchlide reduction' reaction. LPOR, the enzyme that the seedlings of flowering plants use to catalyze Pchlide reduction, is only active in the presence of light, meaning that they cannot produce chlorophyll in the dark. However, plants such as conifers and algae, and photosynthetic bacteria such as R. capsulatus, possess a light-independent (dark-operative) enzyme called DPOR, which consists of two components, an L protein and an NB protein.
"To understand better how DPOR catalyzes Pchlide reduction, we collaborated with X-ray crystallographers, including Genji Kurisu, at Osaka University, to solve the structure of the catalytic component, the NB protein, of DPOR from R. capsulatus," explains Fujita. They analyzed both Pchlide-bound and -unbound states of the NB protein (Fig. 1), revealing that DPOR is similar to nitrogenase, the enzyme involved in biological nitrogen fixation. The L protein of DPOR is known to be structurally related to the nitrogenase Fe protein, which contains iron, while the NB protein is similar to nitrogenase MoFe protein, which contains both iron and molybdenum.
"Surprisingly, we found that the spatial arrangement of the elements for electron transfer in the NB protein and the MoFe protein is conserved very well," says Fujita.
Specifically, they found that each catalytic element of the NB protein contains one Pchlide and one iron-sulfur cluster (NB cluster) coordinated uniquely by one aspartate and three cysteine amino-acid residues. The attachment to aspartate is not necessarily needed for cluster assembly, but is essential for catalytic activity.
Based on their findings, the researchers have proposed a mechanism by which DPOR catalyzes the Pchlide reduction reaction, involving complex chemical and biophysical interactions between the NB protein and bound Pchlide.
Affiliated Researchers
The Nagoya University affiliated researchers mentioned in this highlight are from the GCOE for Advanced Systems Biology: Designing the Biological Function (SYSBIO).
Reference
- Muraki, N., Nomata, J., Ebata, K., Mizoguchi, T., Shiba, T., Tamiaki, H., Kurisu, G. & Fujita, Y. X-ray crystal structure of the light-independent protochlorophyllide reductase. Nature 465, 110-114 (2010). | article