Highlights
Highlights
Nanotechnology opens up for "ultrafast" genetic testing
- Read in Japanese
- ツイート
- 2015/03/12
- Graduate School of Engineering
- Assistant Prof. Takao Yasui
- Prof. Yoshinobu Baba
Prof. Yoshinobu Baba, Assistant Prof. Takao Yasui, and their colleagues at the Graduate School of Engineering, Nagoya University, conducting a joint research with Prof. Tomoji Kawai and his research group at The Institute of Scientific and Industrial Research, Osaka University, newly developed a nanoscaled genetic testing device called "nano-Christmas tree." Using this device, the researchers recorded the fastest deoxyribonucleic acid (DNA) analysis in seconds and made instant genetic testing possible.
This research was published in Scientific Reports on June 11, 2014.→ Nagoya University Press Release
Nowadays, we are more likely to think of ourselves as genes. Nanotechnology is the one that has made it possible.
Constitution, personality, facial expression, and diseases ―are inherited from parents to children: the human body is maintained on the basis of the information stored in genes.
Since the human genome was completely decoded in 2003, the use of genetic information has played a major role in selecting appropriate and optimal therapies adapted to individuals' characteristics (i.e., personalized medicine). Particularly, using genetic testing, administering medical care at the individual level is possible now; for example, identifying genes responsible for various genetic diseases and examining the effects and side effects of a drug prior to its administration.
However, the time required for DNA analysis is currently in the range of several tens of hours. (Please read this article to find out whether the "tens of hours" is fast or slow.) This is because the cells in our body have various quantities of DNA, necessitating a variety of separation conditions appropriate for the various sizes.
Prof. Yoshinobu Baba and his research group at the Graduate School of Engineering, Nagoya University, have developed the technology that makes the genetic testing ultrafast. Their research field is now known as "nanobiodevices," which is an interdisciplinary field between biotechnology and nanotechnology. However, when Prof. Baba started his research, it was the late '80s, when the term nanotechnology had not yet been coined.
By inventing the "inexpensive, fast, and short" genomic testing method, Prof. Baba and his colleagues, today, have become the world's leading researchers in nanobiodevices.
*******
In 1977, DNA sequencing was first established by Frederick Sanger (the Sanger method). Following this, sequencing devices using gel (polymerase chain reaction: PCR) were developed; however, it was believed that it would take 100-300 years to read the whole DNA of one person using the technology of the time.
Thus, the "Human Genome Project" was started as international research collaboration.
In 1996, the first capillary array sequencers were installed in participating laboratories and the human genome was completely mapped for the first time in 2003. It took only 13 years to read the whole DNA of an individual person; in other words, it could be read in 3 months if 300 sequencers were used.
"Still, at this point (in 2001), we needed to achieve 100 million times of efficiency better over the next 30 years."
In 2001, Prof. Baba was already addressing the next step. As the world's population is approximately 7 billion, the speed of reading the human genome in "3 months with 300 sequencers" for each person is too slow. In addition, it would cost 1 billion dollars for a trial at this speed.
Prof. Baba, as a pioneer, insisted that nanotechnology must be used in this field.
His views received some criticisms from other researchers with regard to the challenging aspects of this approach. However, he believed in the necessity of a "grand challenge" to fulfill a radical innovation and established a project that he is conducting with Prof. Tomoji Kawai, Osaka University (NEDO advanced nanobiodevice project).
"Because I knew how much we have achieved so far and what we should do next."
Prof. Baba was confident. He claimed that if we have small goals, we can only accomplish small innovations. Indeed, if we want to face a big challenge, we must not choose a research theme that would be the continuation or extension of what we have been doing.
Usually, projects must produce results within only a couple of years. Thus, for researchers, they have to plan for a desirable future and work and develop concrete strategies backwards connecting the future to the present (backcasting). The fiercely competitive field for innovation is not just limited to Japan but extends worldwide.
"You know, a DNA molecule is also nano sized."
NB: 1 nm = 10-6 mm (i.e., nano means 1/1million milli).
Prof. Baba and Prof. Kawai, his collaborator, started the project "Research and Development of Innovative Nano-biodevices Based on Single-Molecule Analysis" adopted by the Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST) (Nagoya University's FIRST Research Center for Innovative Nanobiodevices).
The research picked up speed.
There are many rivals around the world because the research is important and attractive. Since 2007, the genome-reading speed has dramatically increased (Figure 1). In addition, a trial costs only 1,000 dollars now, compared with about 10 billion dollars in 2001 (Figure 2).
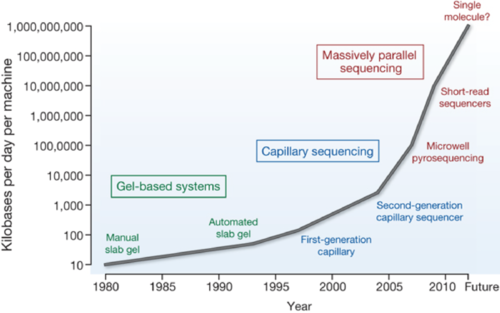 Figure 1. Past 30 years of DNA sequencing speed and its future prediction (the figure cited from:MR Stratton et al. Nature (2009) 458: 719-24 Copyright © 2009, Rights Managed by Nature Publishing Group)
Figure 1. Past 30 years of DNA sequencing speed and its future prediction (the figure cited from:MR Stratton et al. Nature (2009) 458: 719-24 Copyright © 2009, Rights Managed by Nature Publishing Group)
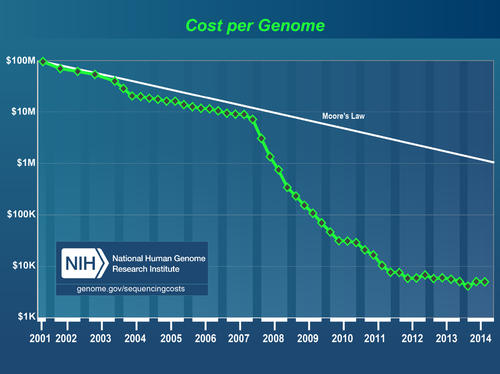 Figure 2. The cost per genome (the figure cited from:NHGRI Genome Sequencing Program (GSP))
Figure 2. The cost per genome (the figure cited from:NHGRI Genome Sequencing Program (GSP))
One of the contributions to this progress is nanopore technology (reported by the United States National Institute of Health (NIH) in 2008.) In fact, the nanopore technology developed in the Kawai laboratory at Osaka University has gathered further hope (Kawai, et al. Nature Nanotechnology (2010) 5: 286-90).
A nanopore, with the pore (approximately 2 nm in diameter) as small as DNA molecules, allows a string of DNA to pass through. Because electrodes are placed in the nanopores, the difference in speed according to the type of DNA bases enables the reading of the DNA base sequence. The speed is 1 base pair (1bp) = 1 millisecond. Because the 3.1 billion bps of human genome can be read in parallel, it is possible to sequence the entire genome in only 1 day.
However, there was a problem. Because the sizes of the pores varied, manual separation of DNA was required after blood collection from patients. Manpower and time were needed for this process, and therefore the automated separation of DNA molecules was an important achievement.
Associate Prof. Noritada Kaji in Prof. Baba's research group has developed a nanopillar structure fabricated of the same size (Figure 3), as the separation technology. In 2004, the nanopillar-based devices successfully separated six different sizes of DNA in only 680 seconds, which was the world's fastest separation technology at that time (Kaji, et.al. Analitical Chemistry (2004) 76: 15-22). Nanopillars are arranged in a zig-zag pattern, and the DNA passes between the pillars.
Figure 3. (A) The schematic description of nanofabrication processes. (B~F) The SEM images of nanopillar structures. The scale bars show 500 nm. (The figure cited from : Anal. Chem. (2004) 76: 15-22. Copyright (2004) American Chemical Society.)
Then, in 2006, Assistant Prof. Takao Yasui, who was in his senior year of university, advanced the principle of separation in terms of the structure used. In the zig-zag alignment, small DNA is sequenced first because big DNA is tumbled by the pillars, whereas in the straight pattern, big DNA is sequenced first.
"Anyway, a three-dimensional structure, resembling a natural structure, like a gel, is the ideal one."
"The nanopillar is a two dimensional structure, which still shows limitations," said Assistant Prof. Yasui. When he was thinking about the possible design of a three-dimensional model, as part of the joint research with Prof. Kawai, the nanowire network was introduced by Associate Prof. Takashi Yanagida, who is currently a professor at Institute for Materials Chemistry and Engineering, Kyushu Universtiy, at The Institute of Science and Industrial Research, Osaka University.
Basically, the diameter of the nanowires is approximately 10 nm, and they grow from the bottom-up and form two-dimensional structures. The nanowires are grown on dispersed gold alone. Therefore, Assistant Prof. Yasui came up with the idea to attach gold on the outer sides of the nanowires, which would allow them to grow in three dimensions (Figure 4).
 Figure 4. The schematic diagram shows the growth process of 3D nanowire. I, II, III are when N=1, 3, 5, respectively. (The figure was cited from : Sakon Rahong et al. Scientific Reports (2014) 4: 5252 Copyright © 2014, Rights Managed by Nature Publishing Group)
Figure 4. The schematic diagram shows the growth process of 3D nanowire. I, II, III are when N=1, 3, 5, respectively. (The figure was cited from : Sakon Rahong et al. Scientific Reports (2014) 4: 5252 Copyright © 2014, Rights Managed by Nature Publishing Group)
Advised by Prof. Yanagida, when the gold catalyst was sputtered under low-vacuum conditions, Assistant Prof. Yasui succeeded in preparing nanowires with gold catalyst on the outer sides. By alternately sputtering the gold catalyst and growing the nanowires, the nanowires were built in three dimensions (Figure 5).
 Figure 5. The SEM images of 3D structure of nanowire. The scale bars show 1 μm(※1 μm=1/1000th of 1 mm). I, II, III refer to N=1, 3, 5 as in the Figure 4. (The figure was cited from : Sakon Rahong et al. Scientific Reports (2014) 4: 5252 Copyright © 2014, Rights Managed by Nature Publishing Group)
Figure 5. The SEM images of 3D structure of nanowire. The scale bars show 1 μm(※1 μm=1/1000th of 1 mm). I, II, III refer to N=1, 3, 5 as in the Figure 4. (The figure was cited from : Sakon Rahong et al. Scientific Reports (2014) 4: 5252 Copyright © 2014, Rights Managed by Nature Publishing Group)
It was Christmas time when the three-dimensional nanowires were completed. Because they look similar to a Christmas tree, Assistant Prof. Yasui and his colleagues call the nanowire structure "nano-Christmas tree."
The nano-Christmas tree functions by entangling DNA in its three-dimensional structure. Various sized DNA (from 100bps to 166kbps, where 1 kbps = 1,000 bps) can be analyzed within 13 seconds. Compared with the separation method used by nanopillars in 2004, as a reference, the nano-Christmas tree can separate the same six types of DNA within 12 seconds: this is 50 times faster than the nanopillar structure.
Here, please see the amazing DNA sequencing speed for yourself (in the movies below).
NB: Don't miss clicking the stop button.
DNA sequencing speed comparison between 5-cycle-growth nanowires (DNA size: 100bps~166kbpsDNA).
【Electric field intensity: 500V/cm】
(The movie was cited from : Sakon Rahong et al. Scientific Reports (2014) 4: 5252 Copyright © 2014, Rights Managed by Nature Publishing Group)
DNA sequencing speed comparison where 38kbps, the DNA size in gel under 300V/cm and in 5-cycle-grown nanowires under 50V/cm.
(The movie was cited from : Sakon Rahong et al. Scientific Reports (2014) 4: 5252 Copyright © 2014, Rights Managed by Nature Publishing Group)
*******
In May 2013, McKinsey & Company selected "next-generation genomics" as one of the "disruptive technologies," mentioning the fact that it allows "inexpensive, fast, and short" genomic testing. By 2025, this technology is expected to produce a market of 0.7-1.6 trillion dollars (McKinsey & Company, May 2013.)
A disruptive technology is one "that could drive truly massive economic transformations and disruptions in the coming years" (cited from McKinsey & Company, May 2013.)
A real innovation requires the change of guidelines for a brand-new value.
―Engineers can change the world.
(Ayako Umemura)
Researchers featured in this article
Dr. Yoshinobu Baba【Professor, Graduate School of Engineering, Nagoya University】
Dr. Baba graduated from the Department of Chemistry, Faculty of Science, Kyushu University, in 1981, and completed his doctorate degree in 1986. Dr. Baba moved to Oita University in 1986 to work as an assistant professor and lecturer. He became a lecturer and associate professor at Kobe Pharmaceutical University in 1990, and a professor at The University of Tokushima in 1997. Since 2004, Dr. Baba is a professor at the Graduate School of Engineering, Nagoya University. Concurrently, he is a director of the FIRST Research Center for Innovative Nanobiodevices, Nagoya University; a director of Nagoya University Synchrotron Radiation Research Center, a professor at Institute of Innovation for Future Society, Nagoya University; a professor at the Graduate School of Medicine, Nagoya University; and an advisor at the Health Research Institute, National Institute of Advanced Industrial Science and Technology (AIST). His specialties are analytical chemistry and nanobioscience.
Prof. Baba has received many awards during his career:
Merck Award (2004);
Division Award of the Chemical Society of Japan (2008); etc.
***
Dr. Baba advances his research by cooperation among industry, academia, and government. Throughout the interview with him, I was thrilled as it felt like I was attending a lecture on how to create innovation.
What can you expect in 2025? Our society and our life itself will be changed by innovation. I'm looking forward to a future that incorporates the great progress achieved by researchers in his laboratory (by AU)
Dr. Takao Yasui【Assistant Professor, Graduate School of Engineering, Nagoya University】
Dr. Yasui graduated from the Department of Chemical and Biological Engineering, School of Engineering, Nagoya University, in 2007. In 2009, he started his research during his doctoral program at the Graduate School of Engineering, Nagoya University, as a Japan Society for the Promotion of Science (JSPS) Doctoral Course student (DC1). In 2011, Dr. Yasui obtained his fast-track doctorate degree in Engineering from Nagoya University, and continued his research as a JSPS postdoctoral fellow. Since 2012, he has been working as an assistant professor at the Graduate School of Engineering, Nagoya University. Concurrently, since 2014, Dr. Yasui has been an assistant program manager at the Impulsing Paradigm Change through Disruptive Technologies Program (ImPACT). His specialties are analytical chemistry, nano-materials, and nano-spatial science.
Assistant Prof. Yasui has received many awards during his career:
The 6th Wakashachi Young Scientists' Award (2012);
The 29th Inoue Research Award for Young Scientists(2013);
Young Innovator Award in Chemistry and Micro-Nano Systems (2014); etc.
***
The day of the interview with Dr. Yasui was a snowy day and the town of Nagoya was completely white (NB: People in Nagoya are not used to snowy streets). Although Dr. Yasui had to leave for Tokyo on his business after the interview, he kindly took me on a tour the laboratory. I enjoyed the tour and there was also a great surprise!
Dr. Yasui appears relaxed; at the same time, he exhibits dynamism and a strong view for innovation. I expect his further research achievements (by AU)
Links
- Nagoya University's FIRST Research Center for Innovative Nanobiodevices HP http://www.nanobio.nagoya-u.ac.jp//e_index.html
- Baba Laboratory HP http://www.apchem.nagoya-u.ac.jp/III-2/baba-ken/index.html
- New Energy and Industrial Technology Development Organization (NEDO) advanced nanobiodevice project http://www.nedo.go.jp/activities/ZZ_00285.html
- Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST) http://www.jsps.go.jp/english/e-first/index.html
- The United States National Institute of Health (NIH) http://www.nih.gov/
- McKinsey & Company
http://www.mckinsey.com/insights/business_technology/disruptive_technologies
- Link to this article (from a search-result)
http://www.sciencedirect.com/science/article/pii/S2211124714008602
Sakon Rahong, Takao Yasui, Takeshi Yanagida, Kazuki Nagashima, Masaki Kanai, Annop Klamchuen, Gang Meng, Yong He, Fuwei Zhuge, Noritada Kaji, Tomoji Kawai & Yoshinobu Baba
Ultrafast and Wide Range Analysis of DNA Molecules Using Rigid Network Structure of Solid Nanowires.
Scientific Reports 4: 5252 (2014).
(First published on June 11, 2014; doi:10.1038/srep05252)
- Link to related article1 (from a search-result)
http://www.nature.com/nnano/journal/v5/n4/full/nnano.2010.42.html
Makusu Tsutsui, Masateru Taniguchi, Kazumichi Yokota & Tomoji Kawai
Identifying single nucleotides by tunnelling current.
Nature Nanotechnology 5: 286 (2010).
(First published on March 21, 2010; doi: 10.1038/nnano.2010.42)
- Link to related article2 (from a search-result)
http://pubs.acs.org/doi/abs/10.1021/ac030303m
Noritada Kaji, Yojiro Tezuka, Yuzuru Takamura, Masanori Ueda, Takahiro Nishimoto, Hiroaki Nakanishi, Yasuhiro Horiike, and Yoshinobu Baba
Separation of Long DNA Molecules by Quartz Nanopillar Chips under a Direct Current Electric Field.
Anal. Chem. 76: 15 (2004).
(First published on November 26, 2003; doi:10.1021/ac030303m)
NU Research
(English)




